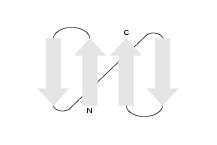
By loop lengths we mean the lengths of the segments of the protein
between the strands in the given sheet. Note that this can include
helical structures, and even strands (if they are in different
sheets). We define two classes of length type: long, if the segment
between two strands in the given sheet is more than 10 residues, and
short if the segment has ten or less residues. Long "loops" are
labelled with a 1 on the form, short loops with a 0. We follow the
strands along the backbone of the protein from the N-terminus to the
C-terminus. As an example, we consider protein L, which has the
following sheet motif:

The amino acid sequence and the secondary structure elements (according to dssp) are shown below. The dash represents everything that is not in a strand or a helix.
ENKEETPETPETDSEEEVTIKANLIFANGSTQTAEFKGTFEKATSEAYAYADTLKKDNGEYTVDVADKGYTLNIKFAG -----------------SSSSSSSS------SSSSSSS--HHHHHHHHHHHH--------SSS-------SSSSSS--
According to dssp, there are 6 residues between strand one and two, 22
residues between strand two and three, and 7 residues between strand
three and four. The loop lengths therefore are short-long-short,
indicated as 0-1-0 on the form.